Restriction endonucleases are enzymes that cut DNA at specific sequences, usually 4 to 6 base pairs in length. For example, BamHI cuts only at DNA sequences GGATCC.
Since genes have characteristic DNA sequences, the sites at which different restriction enzymes cut a gene are unique to that gene. A diagram showing the location of the restriction enzyme cleavage sites in a gene, or larger piece of DNA, is known as a restriction map.
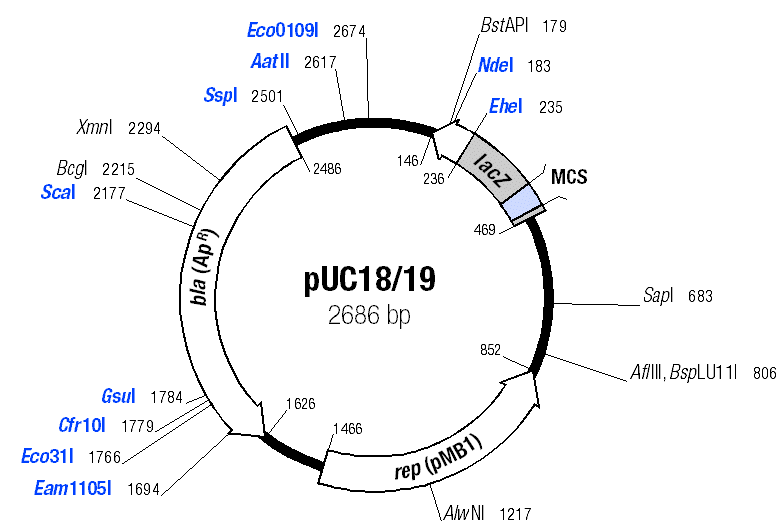
In this experiment you have isolated two different plasmid DNA molecules, pUC18-HIS3 and pUC18-URA3. Since they have different gene components they will differ in their restriction maps, and you will be able to identify them by the sizes of the DNA fragments produced by restriction enzyme digestion.
You can put together the restriction map of each plasmid, and predict the DNA fragment sizes, by assembling the plasmids from sequences you will retrieve from databases.